 |
atom_comp
Brian Sykes Lab
|
 |
Version: 2.0.1 - Mar 7 / 2014
Download Here
Purpose: Atom comparison and energy calculations for an ensemble pdb file
Reference
Lindhout, D.A., Boyko, R.F., Corson, D.C., Li, M.X. and Sykes, B.D. (2005)
The role of electrostatics in the
interaction of the inhibitory region of troponin-I with troponin-C.
Biochemistry 44(45):14750-14759.
Software
Release Notes
Installation
- First, before downloading the software, make sure you have "wish"
in your path. Type 'which wish' in a terminal window and it should come
back with a location
(eg, /usr/bin/wish). If you don't have it, you may need to install
tcl/tk on your machine first.
- Next, download the software from
www.bionmr.ualberta.ca/download/atom_comp
- untar it
> tar xvf atom_comp*.tar
- Copy the atom_comp script to a public place or your $HOME/bin directory.
> cd atom_comp/src/bin
> cp atom_comp /usr/local/bin
Example
- Make sure you have an ensemble pdb file. In this example we have 30
models. Use spaces or tabs to delimit the fields. Each line
contains one atom pair for comparison.
- Place protein atoms in a file
# Protein atoms
LYS 90 A NZ 1.602E-19
LYS 92 A NZ 1.602E-19
ARG 102 A CZ 1.602E-19
LYS 106 A NZ 1.602E-19
LYS 118 A NZ 1.602E-19
LYS 138 A NZ 1.602E-19
LYS 142 A NZ 1.602E-19
ARG 147 A CZ 1.602E-19
LYS 158 A NZ 1.602E-19
GLU 94 A CD -1.602E-19
GLU 95 A CD -1.602E-19
GLU 96 A CD -1.602E-19
ASP 99 A CG -1.602E-19
...
- Place peptide atoms in a file
# Peptide File
LYS 139 B NZ 0
ARG 140 B CZ 0
THR 142 B OG1 0
ARG 144 B CZ 0
ARG 145 B CZ 0
ARG 140 B CZ 0
ARG 147 B CZ 0
LYS 137 B NZ 0
ASP 133 B CG 0
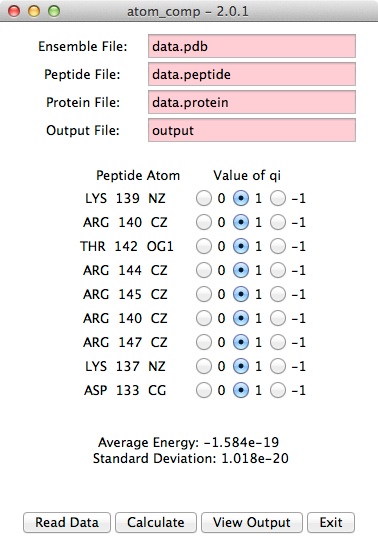
- Within a terminal window type "atom_comp". You should get the
following atom_comp gui. Next, enter your 3 input files and an output file.
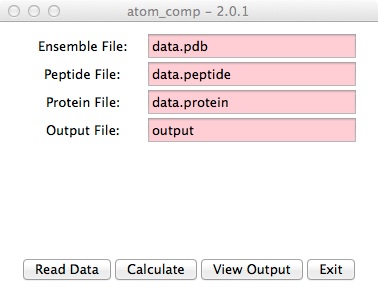
- Click the "Read Data" button.
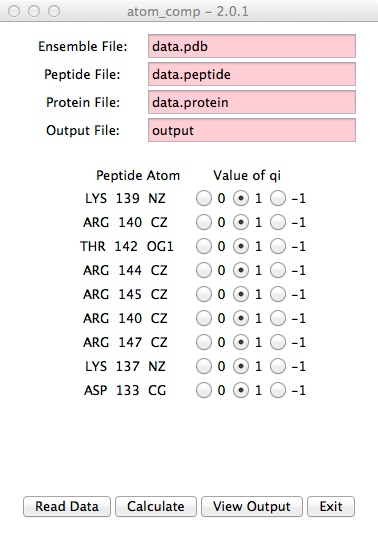
- Select the atoms to use in energy calculations.
Click the "Calculate" button.
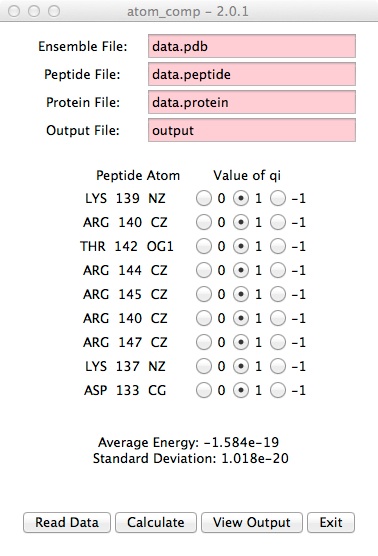
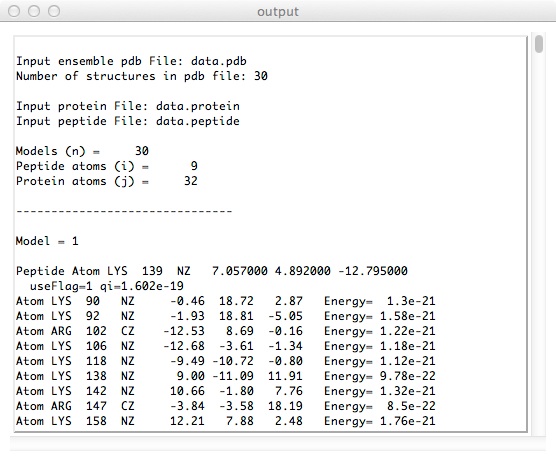
- The output will look similar to this.
 Back to main software page
Back to main software page
This file last updated:
Questions to:
bionmrwebmaster@biochem.ualberta.ca








