 |
ppscan Brian Sykes Lab |
 |
 |
ppscan Brian Sykes Lab |
 |
Version: 1.0.1 - Mar 5 / 2014
Download Here
Purpose: Allow the user to extract parameter values from a vnmrj procpar file for all the fids located in a directory. Output the resulting table to be read and analyzed with Excel. We use this script in our lab to see how shims have changed for various related vnmrj experiments.
Download the software from www.bionmr.ualberta.ca/download/ppscan and untar it. > tar xvf ppscan*.tar > cd ppscan/src/bin Copy the ppscan script to a public place or your $HOME/bin directory. > cp ppscan /usr/local/bin
Start a terminal window and issue a command in the following format: Usage: ppscan input_fid_dir parm1 [parm2] [parm3]... > output.xls where input_fid_dir - A directory containing all the fid data for analysis parm1 - the first parameter to extract from the procpar file parm2 - the (optional)second parameter to extract from the procpar file parm3 - the (optional)third parameter to extract from the procpar file ... > output.xls - direct the results to this file. The xls suffix allows excel to read it.
# Example command > ./ppscan $HOME/vnmrsys/data x1 x2 y1 y2 z0 Also see test/test.run for an example of how one can run the script and what the output should look like. Here is a quick example after importing the results into excel.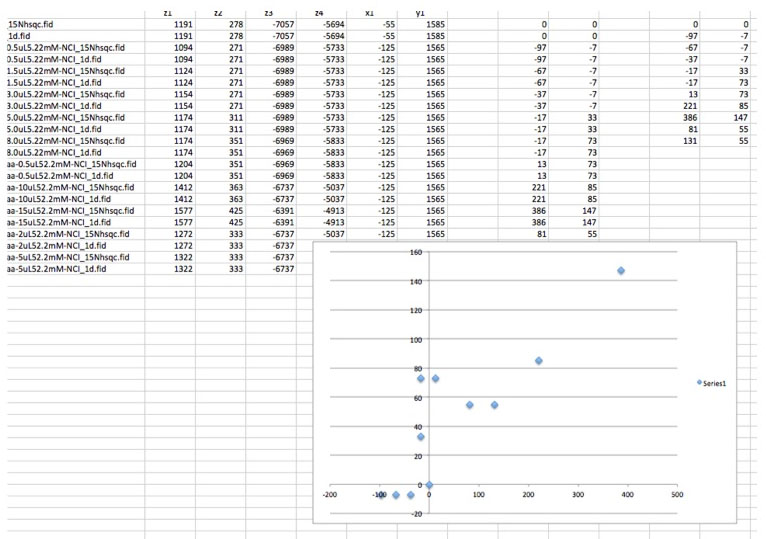
This file last updated: Questions to: bionmrwebmaster@biochem.ualberta.ca