- peakId
- peaklist
- atom name
In the example that follows, the user is designating peakId 2 of the reference spectra as being folded in the Nitrogen dimension (just pretend that it is). The peakId was either typed by the user or set by one of the functions in the smartnotebook "Reference Id" menu.
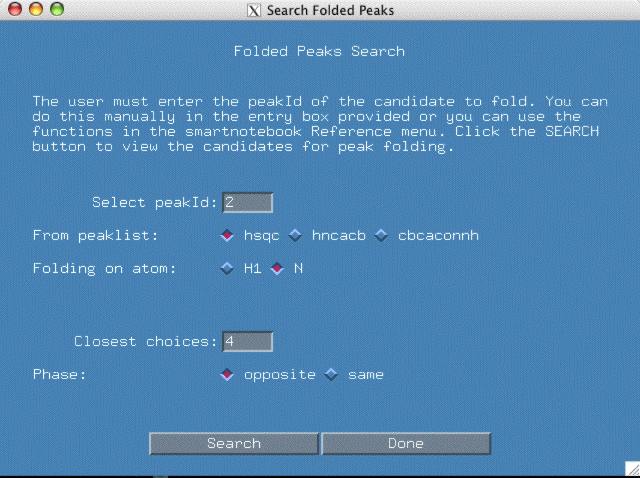
By designating the folded peak above, the software needs to search the other peaklists for corresponding peaks that will also need to be folded.
The "Closest Choices" field means the closest 'n' choices to your selected peak in each peaklist will be displayed, usually 4 is a high enough number. If you select "opposite" phase, the intensity of the folded peak will be multiplied by -1.
At this point, one can click the "Search" button. You should see a new popup window similar to the one below.
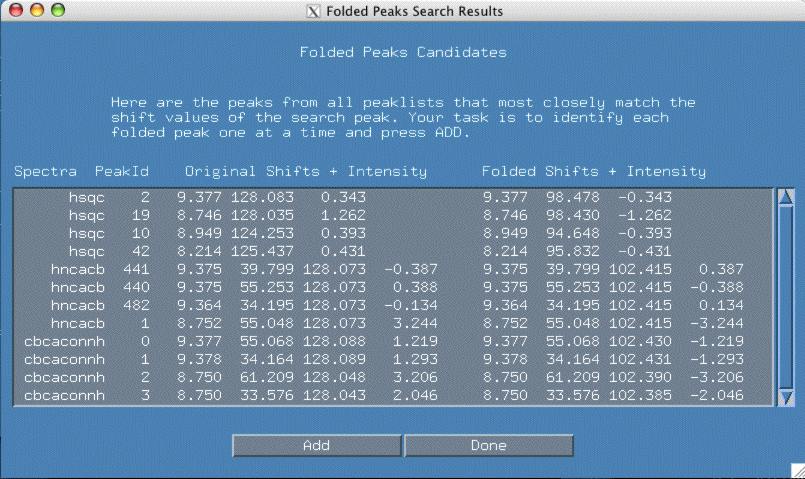
It is now your job to select each peak which you believe is folded and click "Add" which adds it to your current folded peak list. Depending upon the spectra type, multiple peaks may be folded in the same spectra. Click the "Done" button when there are no more peaks to add.
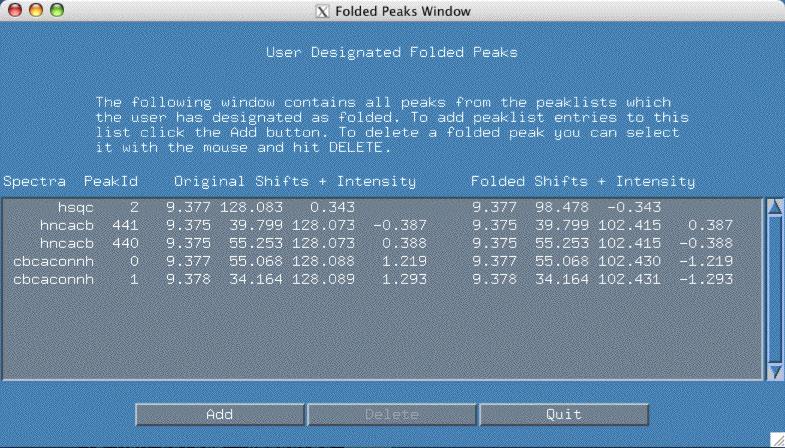
If you go back to the original folded popup window as shown in the figure above, you can see how smartnotebook shows you the exact nature of the adjustment to your peakpick files. In the above example, columns 3-6 show you the original peak, columns 7-10 show you the adjusted folded peak. If at any time you realize a mistake has been made, select the offending peak line and hit the "Delete" button.
 Smartnotebook Home
Smartnotebook Home