This example demonstrates the use of smartnotebook with the hn(co)ca/hnca experiment data. Thanks to Pierre-Yves Savard in Stéphane Gagné's lab for providing data for this project.
> cd snb-v5.1.3-examples2 > cd hnco-yves If you are on a linux machine, you will need to edit each par file and make sure byteorder is set to 1. For the other platforms, this parameter is set to 0 (as far as I know). > snbview
When nmrview comes up, type "snb" at the nmrview console.

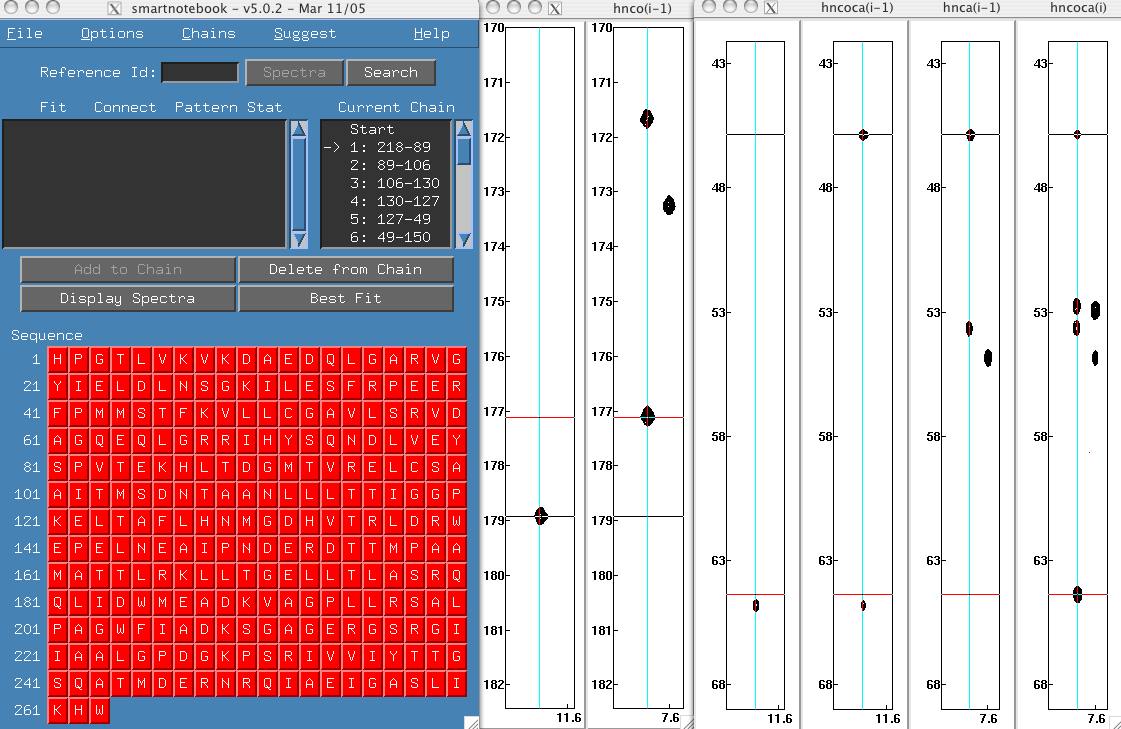
To configure snb you need to select the "hnca" and "hnco" buttons in the "Initialization procedure (2 of 6)". Complete the initialization process as in previous examples, then move the snb tableau and spectra to convenient locations on your screen. Here it is convenient to make a larger snb window. The nmrview console indicates that 9345 connections were found, now that is a lot of possibilities to decipher!
If you click the chains menu, you will see that there is a large chain created however no possible assignment is displayed in the sequence window. Click on the "best fit" button and you will see that Gly 3.hn has a very unusual shift. We should really edit our shifts constraints file (snb.out/shifts) which was generated from BMRB statistics to account for this unusual shift. However, in this case we are going to take the easy way out and move the stddev slider bar from 3.0 to 4.75.
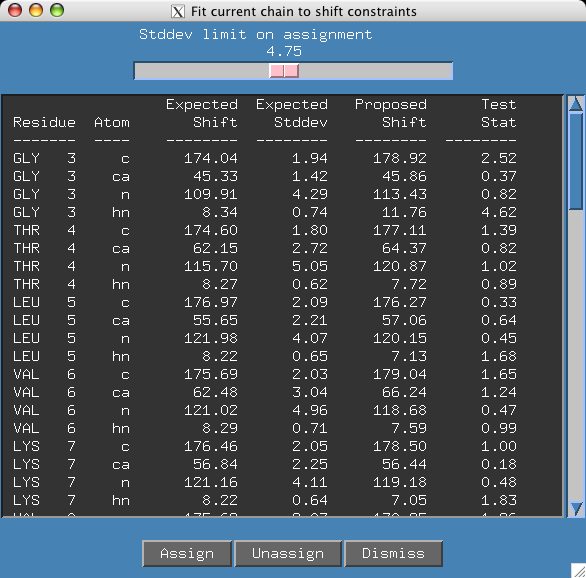
It appears we have an unambiguous assignment but if we move the slider a bit further to 5.10 a second assignment appears. If you click on the "G171" residue in the sequence panel it is easy to see that several shifts do not have good agreement between proposed and expected values.
It is very difficult to assign such a large protein without having Cb to help distinguish the amino acids. To set the stddev slider at 4.75 will also make the task more difficult. Certainly in these types of experiments working with a tighter shift constraints file would also help.
Your task for this example is 'walk' through the chain to see how easy/difficult it was to come up with the correct choices. To do this, you can go to the chains menu and select "Start new Chain". It would also be a good idea to click on the first separator line which turns the menu into a window. Now you can see the chain you are going to re-create.
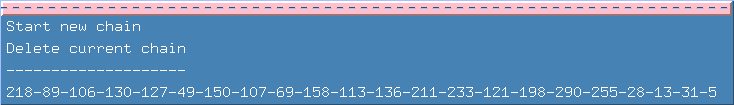
Enter 218 in the "Reference Id" field and press return. Here the correct choice is fifth on the list. Select it and click "add to chain". Click "End" in the Current Chain panel and this time our correct choice is first. Feel free to go through the entire chain, trying some of the incorrect answers and displaying the spectra. Remember that you have the "Delete from chain" button that will allow you to backtrack.
This file last updated: Questions to: bionmrwebmaster@biochem.ualberta.ca