Smartnotebook Example 6: hsqc-suggest
This example shows how to import suggested chains or assignment information
into smartnotebook. Other software would come up with proposed assignments,
then the user could verify that these assignments correctly match the
data. It is interesting work for those who like the smartnotebook
interface and also want the good results from other software.
Start smartnotebook from within the hsqc-suggest directory by typing the
name of the startup script.
> cd hsqc-suggest
> snbview
The nmrview software should have started. If it did not, perhaps you have
not properly set up your "snbview" alias as mentioned in the
" installation page.
Next type "smartnotebook" in the nmrview console window (or "snb" for short).
> snb
After a short wait, you should now see the spectral strips, reference spectra
and smartnotebook main window displayed on your screen.
- Click on the "Suggest" menu item and click "Suggest Chains". You will
get a popup window which asks for the name of a file in either
ppm.out or
chains format.
- Let's import a chains format file first. Click "chains" and enter
the name "suggest.chains". Press execute.
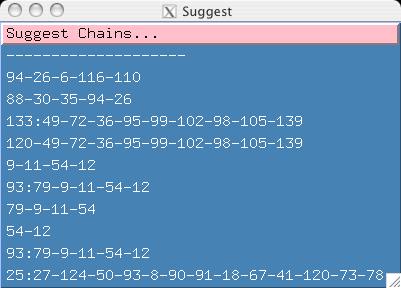
- Now when you select the "Suggest" menu, you can see that all the
suggested chains in the file were read into smartnotebook.
- This time we are going to read in a ppm.out formatted shifts file and
create chains based on assignments of consecutive amino acids. Click on
"Suggest Chains" again and enter the format and filename fields as
specified below.
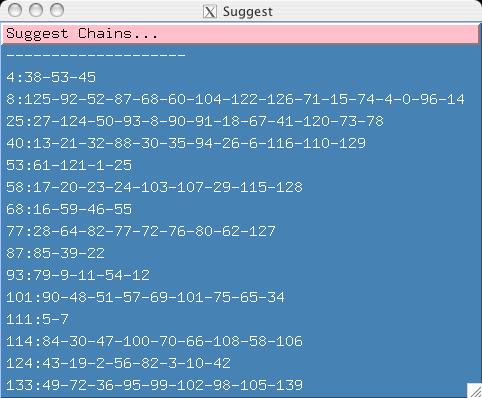
- Notice that we now have a new set of suggested chains for the user to
examine.
- A suggested chain is not part of the user's chain list until the
user clicks on it. Try doing this with the first chain in the suggested
list. The chain becomes the current chain, and if the user doesn't like it
then he/she should delete it.
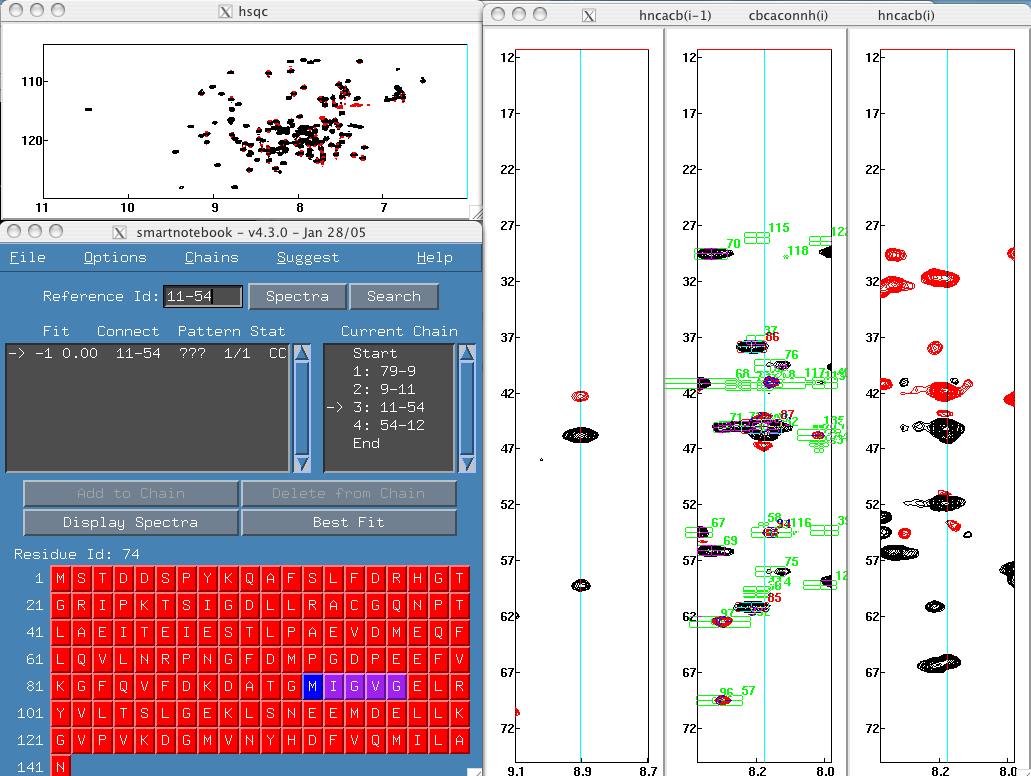
- If a suggested chain is missing connectivity information, the software
designates a connectivity pattern of "???". Therefore the spectroscopist
will know that the current peakpick data does not support the connectivity
and will have to investigate why. Click on the 79-9-11-54-12 chain to
view an example. To display the 11-54 spectra, type 11-54 in the Reference Id
field or click on the 11-54 entry in the chains panel.
When we turn on peak labelling (Options menu), we can see that this is a
valid connectivity as peak overlap caused us to miss peakpicks.
Smartnotebook 4.0 and higher does not plan to support the older versions of snb
where the making of chains from assignments was via the reference xpk files.
The ppm.out format is a simple and broad way to summarize assignment
information and I think it is better in the long run. I think it has to
be the job of snb to figure out how to
handle assignment files with partial assignments and/or without
corresponding peakpick data. Your opinions regarding this topic are welcome.
 Smartnotebook Examples
Smartnotebook Examples
This file last updated:
Questions to:
bionmrwebmaster@biochem.ualberta.ca











